MitoGAN
Project Timeline: Feb 2023 - May 2023 (~4 months)
Skills Used: PyTorch, OpenCV, Generative Adversarial Networks (GANs), Image Processing, Edge Detection (Laplace Filtering), Denoising (Non-local Means), Thresholding (Otsu’s)
Mentors: Gav Sturm, Wallace Marshall
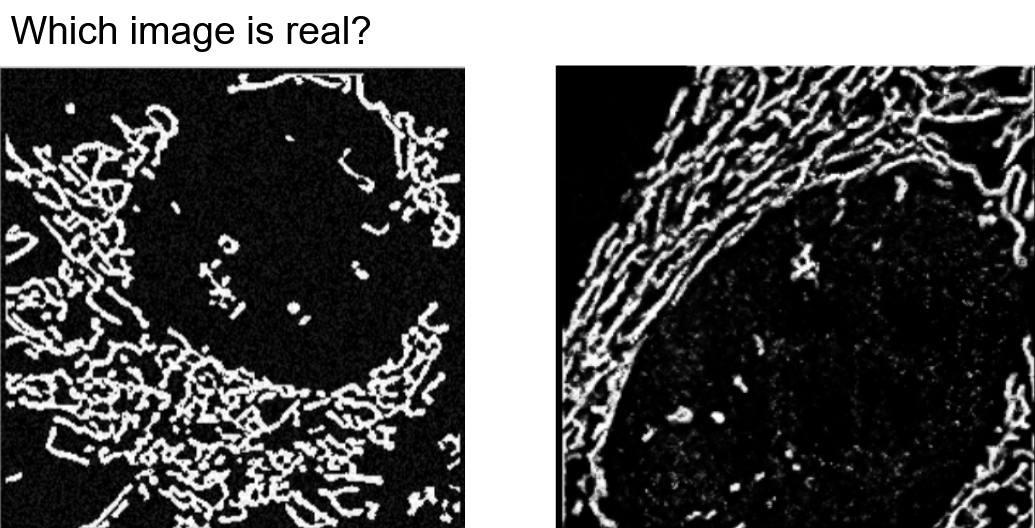
Context:
“Mitochondria are not just beans 🤬!”
— Gav Sturm
Mitochondria are organelles with remarkably complex biophysical structures that have poorly understood mechanical behaviors. These subcellular structures have dynamic morphologies that adapt to the many intricate biological processes within the cell. They stretch, elongate, shrink, bend, and even “bead” almost as if they were cells themselves! I joined Gav Sturm for a rotation project in order to help better understand the remarkable dynamics of mitochondria.
Gav ultimately wanted to capture a biophysical model of mitochondrial dynamics in silico. We approached the problem using a data-driven heuristic. In the early stages of the work, Gav researched, developed, and simulated physics-based parametrized models. Naturally, the next step was to identify the latent parameters underlying the dynamics of mitochondria. I joined during the next stages of the work. Gav and I brainstormed approaches for learning the distribution of features most relevant for recapitulating dynamics, and we deduced that exploring generative machine learning methods seemed the most relevant for this application.
Problem:
We were dealing with largely unstructured data in the form of fluorescence microscopy videos. Additionally, our dataset was rather limited at the time as there were only about 20 videos that added to only about 600 images to use. Lastly, the images required some processing in order to standardize the outputs across the samples (in case there was any drift/distribution shift over time). Processing the samples down may also allow for more sample efficient training which is important in small sample regimes. Summarily, our problem statement boiled down to:
- Data in the form of fluorescence microscopy videos of labeled mitochondria.
- Small sample regime.
- Standardization of samples to account for feature distribution shift and small sample regime.
Solution:
Prior to choosing and training a model, the samples needed to be standardized. I built an image processing pipeline to account for drift across samples and reduce the dimensionality of the input features (reduced dimensionality allows for more sample efficient training). The pipeline is summarized in the following figure:
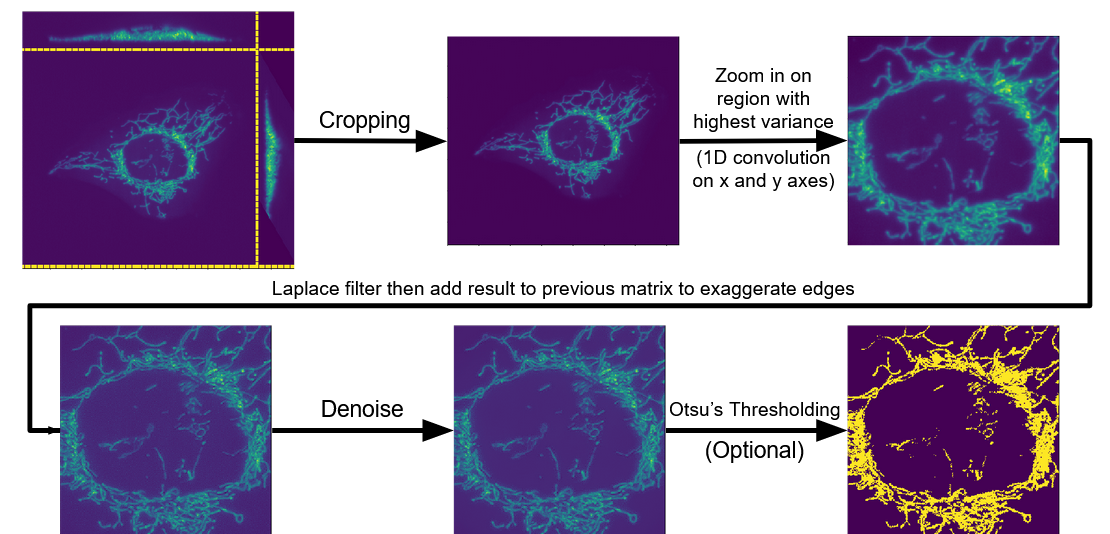
Firstly, sequence modeling was ruled out because
- there were not enough videos (only ~20) to be used as samples and
- using each frame (image) from the videos as its own samples simplified the problem statement.
Secondly, we wanted to use a generative model in order to
- learn the feature distributions from the samples and
- use the synthetic data generated from the models for training more models.
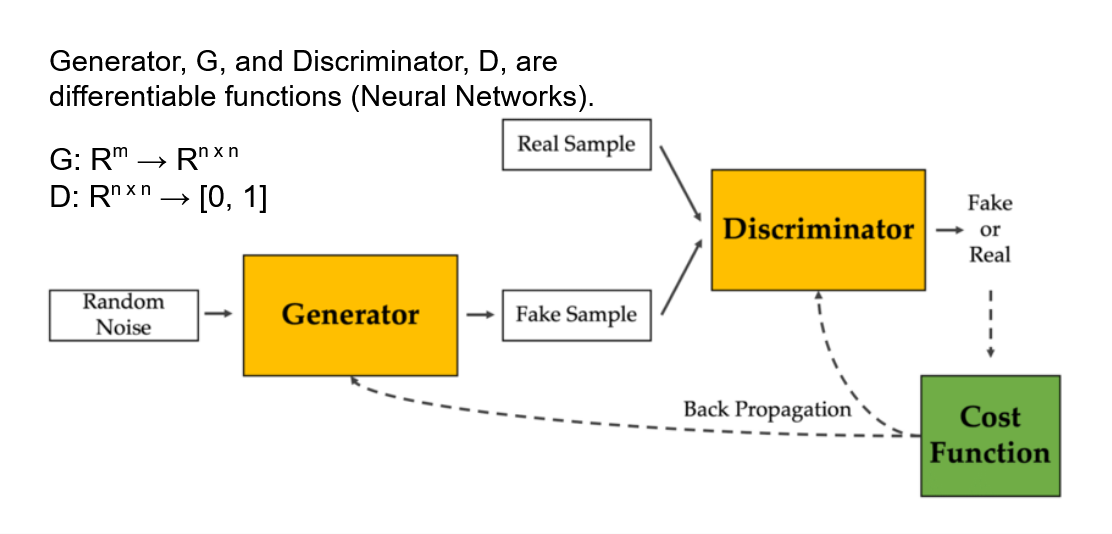
We opted to use Generative Adversarial Networks (GANs) since the generator model is capable of learning a distribution of the input features and can be used to generate synthetic data. Additionally, we can interpret the discriminator model to see what features it found most predictive for classifying between real and fake images. In other words, the GAN architecture offers both the benefits of learning the features’ distributions (generative) and interpreting which geometrical features are most important to classifying mitochondria (discriminative).
Results:
To resummarize the objectives, we used GANs in order to
- capture the distribution of input features within a model,
- generate synthetic data for training subsequent models, and
- interpret the most salient geometric features for identifying mitochondria.
During the rotation, I was able to complete a first pass at 1 and 2 but not 3. I processed and prepared training and testing data for training and evaluating the GAN. The results are as follows:
- Original dataset: ∼ 600 images; Generated: ∼ 30000; 50x generated
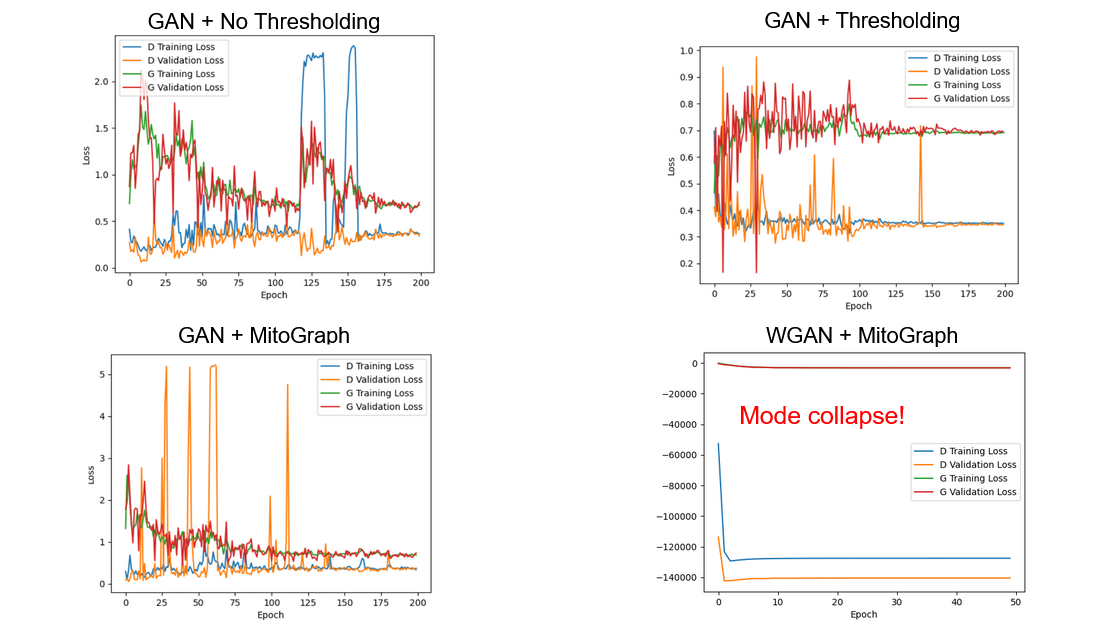
- Tested different preprocessing combinations and GAN architectures to identify stable models with lowest collective losses:
Discussion
Notably, GANs are quite unstable. Additionally, there is a chance they may not converge if not initialized correctly (e.g. as shown in the Wasserstein GAN (WGAN) loss curve).
There are a couple of further analyses that I did not have the chance to conduct:
- The discriminator/classifier still needs to be analyzed for identifying salient geometric features.
- Though many images were generated, the effective number of unique images is likely lower since generated images collapsed into several modes. This is likely due to low diversity of cells imaged in the videos. More training data is needed. (When is it not needed? Lol.)
- These days, there might be more powerful and interpretable methods than using a GAN architecture, such as other self-supervised learning approaches.

